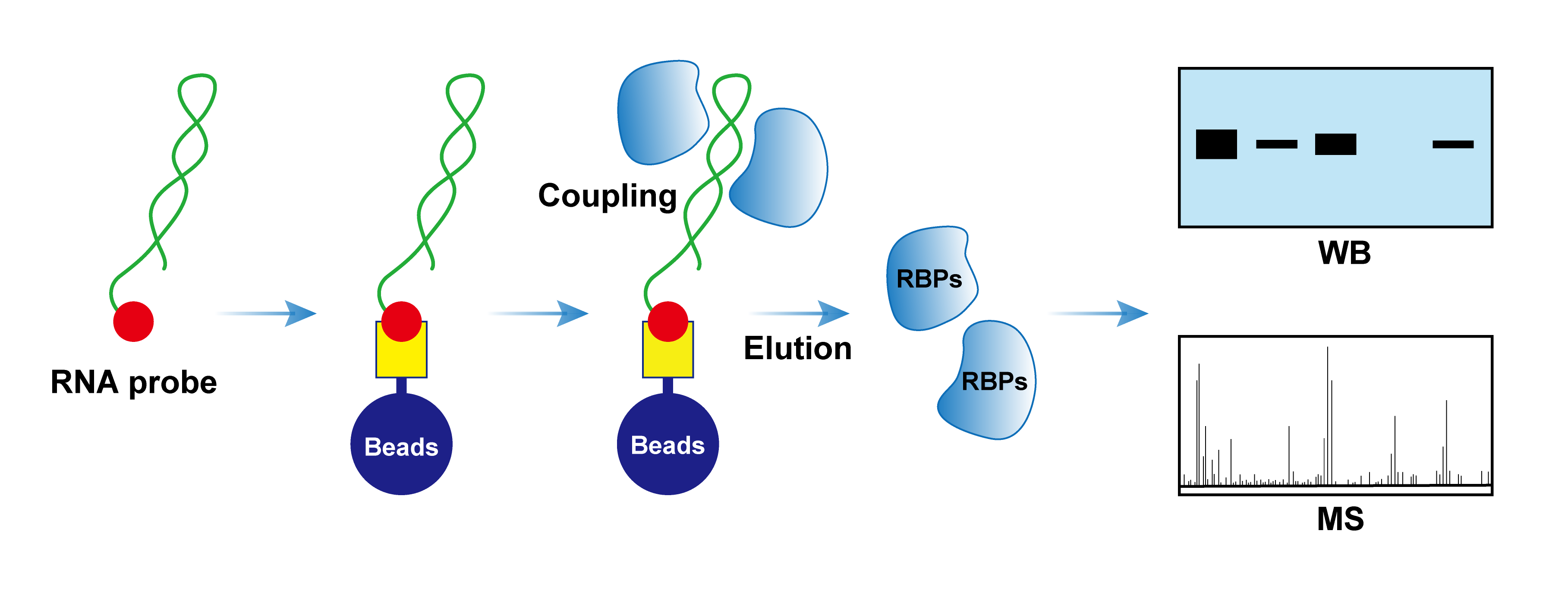
RNA pull-down is one of the main experimental methods for detecting interactions between RNA-binding proteins and RNA. This technique uses in vitro synthesized biotin-labeled RNA to mimic natural RNA molecules as probes, which are incubated with cell lysates. The probes form RNA-protein complexes with proteins in the lysates, which can be specifically bound and captured by streptavidin magnetic beads, thereby achieving capture and enrichment of RNA-binding proteins (RBPs).
Product Features
1. Convenience: The kit contains complete components, including the full set of reagents for the pull-down experiment;
2. Speed: Can be completed in 2.5 hours;
3. Stability: High capture efficiency, stable experimental system, and high reproducibility.
Product applications
● Validation or screening of target RNA-binding proteins;
● Suitable for long, linear RNA probes (mimicking target RNA sequences) to directly pull down interacting proteins.
Procedure
Results Display
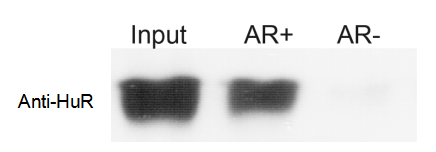
Figure 1.AR+ probe specifically captures HuR protein
Case Analysis
RNA pulldown assay involves biotin-labeling the RNA, incubating it with cell lysate to form RNA–protein complexes, then isolating the complexes to obtain the proteins, which are subsequently detected by WB or mass spectrometry.
We know,
RIP is a technique that studies protein–RNA interactions starting from the protein, using a known protein to verify RNA molecules that may interact; RNA pull-down, on the other hand, is a technique that studies protein–RNA interactions starting from the RNA, using a known RNA to pull down proteins and infer proteins that may bind the known RNA. Therefore, RIP and RNA pull-down can validate interactions bidirectionally, more accurately reflecting RNA–protein interactions.
Case1

Gastric cancer is the fourth most commonly diagnosed cancer worldwide and the third leading cause of cancer-related death globally. Because the pathological mechanisms driving gastric cancer progression are not yet fully understood, more research is needed to discover and develop effective diagnostic biomarkers and therapeutic targets for gastric cancer. In this paper, the authors discovered and named a novel
lncRNA_GClnc1. It acts as a scaffold lncRNA to recruit the proteins WDR5 and KAT2A and activates transcription of the target gene SOD2, ultimately promoting gastric cancer progression.
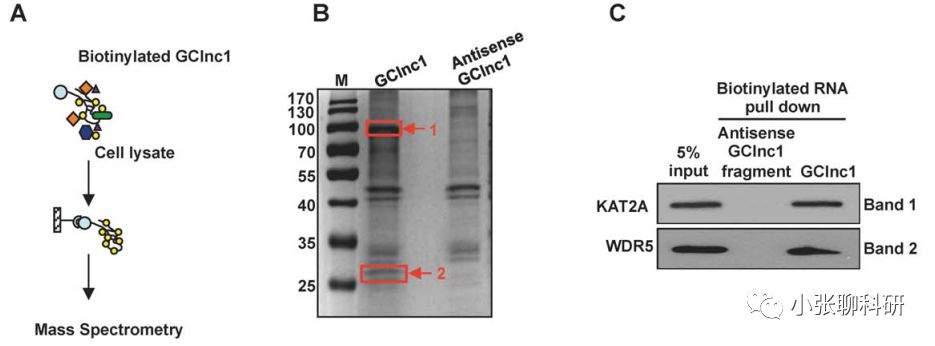
图
1. GClnc1 RNA pull-down result image
In this study, in order to explore
the mechanism by which GClnc1 promotes gastric cancer, the authors used RNA pull-down analysis to identify intracellular GClnc1 binding factors (Fig-1A). Two specific bands were present in the GClnc1 pull-down samples. The authors identified 94 and 101 potential binding proteins (Fig-1B). Western blotting showed that only WDR5 and KAT2A specifically bind GClnc1 (Fig-1C). The data suggest that these two proteins may form a complex to interact with GClnc1.[1]
Case2

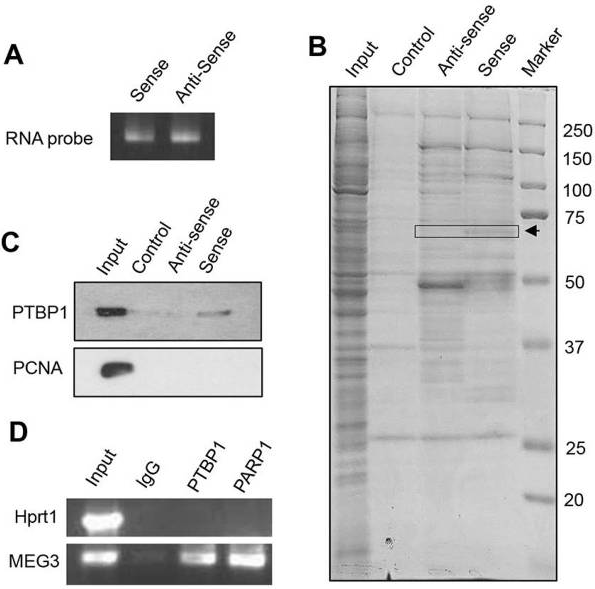
图
1 Pulldown & RIP reveal interaction between PTBP1 protein and MEG3 RNA
(A) Sense and antisense (negative control) biotin-labeled MEG3 RNA synthesized by in vitro transcription
(B) In vitro RNA pull-down assay. HEK293T cell lysates were incubated with biotin-labeled oligonucleotides. After pull-down, proteins were subjected to SDS-PAGE and stained with Coomassie Blue. The bands indicated by arrows were analyzed by mass spectrometry. (C) Western blot to determine the specific interaction of sense MEG3
RNA with PTBP1 protein, but not with PCNA protein (negative control). (D) RNA immunoprecipitation (RIP) showing in vivo interaction of PTBP1 with MEG3
RNA in HEK293T cells. Protein–RNA complexes immunoprecipitated with anti-PTBP1 or IgG were analyzed by RT-PCR using primers specific for MEG3 or Hprt1 (negative control).
In this study, first through
RNA pulldown and subsequent mass spectrometry experiments identified the protein PTBP1 as binding to lncRNA MEG3, and this interaction between lncRNA MEG3 and PTBP1 was further validated by RIP using an antibody against PTBP1.
Case analysis references:
[1] Tian-Tian Sun, Jie He, Qian Liang, Lin-Lin Ren, Ting-Ting Yan, Ta-Chung Yu, Jia-Yin Tang, Yu-Jie Bao, Ye Hu, Yanwei Lin, Danfeng Sun, Ying-Xuan Chen, Jie Hong, Haoyan Chen, Weiping Zou and Jing-Yuan Fang. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Dis. July 2016
[2]
Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017 Feb;65(2)
Customer Article
[1] IF8.469 Linc-ROR facilitates progression and angiogenesis of hepatocellular carcinoma by modulating DEPDC1 expression. Cell Death & Disease
[2] IF6.684 Inhibition of miR-185-3p Confers Erlotinib Resistance Through Upregulation of PFKL/MET in Lung Cancers. Frontiers in Cell and Developmental Biology
[3] IF6.244 circDNMT1 Promotes Malignant Progression of Gastric Cancer Through Targeting miR-576-3p/Hypoxia Inducible Factor-1 Alpha Axis. Frontiers in Oncology
[4] IF5.555
Long noncoding RNA X-inactive-specific transcript promotes the secretion of inflammatory cytokines in LPS-stimulated astrocyte cells via sponging miR-29c-3p and regulating Nuclear Factor of Activated T cells 5 expression. Frontiers in Endocrinology
[5] IF5.502 Silencing of LINC01963 enhances the chemosensitivity of prostate cancer cells to docetaxel by targeting the miR-216b-5p/TrkB axis. LABORATORY INVESTIGATION
[6] IF5.458 Increased LDL receptor by SREBP2 or SREBP2-induced lncRNA LDLR-AS promotes triglyceride accumulation in fish. iScience
[7] IF5.241 The lncRNA MIAT regulates CPT-1a mediated cardiac hypertrophy through m6A RNA methylation reading protein Ythdf2. Cell Death Discovery
[8] IF5.228 C/EBPα promotes porcine pre-adipocyte proliferation and differentiation via mediating MSTRG.12568.2/FOXO3 trans-activation for STYX. BIOCHIMICA ET BIOPHYSICA ACTA-MOLECULAR AND CELL BIOLOGY OF LIPIDS
[9] IF4.599 CircDTL Functions as an Oncogene and Regulates Both Apoptosis and Ferroptosis in Non-small Cell Lung Cancer Cells. Frontiers in Genetics
[10] IF4.36 Circ_0001667 knockdown blocks cancer progression and attenuates adriamycin resistance by depleting NCOA3 via releasing miR-4458 in breast cancer. DRUG DEVELOPMENT RESEARCH
[11] IF3.575 Knockdown of circBFAR inhibits proliferation and glycolysis in gastric cancer by sponging miR-513a-3p/hexokinase 2 axis. BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS
[12] IF2.952
Circular RNA OMA1 regulates the progression of breast cancer via modulation of the miR-1276/SIRT4 axis. Molecular Medicine Reports
[13] IF2.626 Regulation of circADAMTS6-miR-324-5p-PIK3R3 ceRNA pathway may be a novel mechanism of IL-1β-induced osteoarthritic chondrocytes. JOURNAL OF BONE AND MINERAL METABOLISM
[14] IF2.57 Long Non-Coding RNA GABPB1-AS1 Augments Malignancy of Glioma Cells by Sequestering MicroRNA-330 and Reinforcing the ZNF367/Cell Cycle Signaling Pathway. Neuropsychiatric Disease and Treatment
[15] IF2.192 Silencing of Circ_0135889 Restrains Proliferation and Tumorigenicity of Human Neuroblastoma Cells. JOURNAL OF SURGICAL RESEARCH
Q & A
1. Can this pull-down kit be used for circRNA?
Currently the company’s
RNA-Protein pulldown kit is suitable for linear RNA; for circRNA one can consider tag-based methods. The official WeChat account article “New-generation circRNA pulldown research methods” provides a detailed introduction.
2. Can pulldown products detect interacting RNA? Can RNA be extracted using Trizol?
In the current kit system, the product separation system primarily targets proteins and does not involve RNA.RNA extraction and detection.
3. Is the target probe a sense strand probe or an antisense strand?
The target probe is a sense strand probe; the negative control is its antisense strand.
4. What is the recommended amount of probe to use?
Recommended Amount of Probe50 pmol per reaction, or adjust the amount according to the specific experimental conditions.
5. About Biotin-Labeled Probes
The biotin label applied in this kit
RNA probes are intended to mimic native RNA molecules; in theory the more complete the sequence coverage the better. They can be obtained by direct synthesis or by in vitro transcription. However, sequences that are too long (greater than 2 kb) are difficult to obtain, so regions with expected protein binding can be prioritized.
6. How to determine whether there is a problem with the experimental operation?
It is recommended to include a positive control during the experiment; the kit contains a positive control probe and
HuR antibody; HuR protein is present in almost all cell samples and can be pulled down by the positive probe with strong binding. The WB result for positive-control HuR can be used to determine whether the experimental operation has issues; the negative control can be used optionally.
7. The loading buffer in the pull-down kit is rather pale and different from the one usually used; is this normal?
Our company
The loading buffer in the pulldown kit is indeed somewhat paler. This is mainly because an excessive amount of loading buffer can affect nucleic acid electrophoresis results to some extent, so it is sufficient as long as the bands are visible.
8. What are the similarities and differences between RAP (RNA antisense purification) and RNA–protein pulldown?
Similarities: Both are probe-based pull-downsRNA-binding complexes.
Differences:
1)
RAP essentially uses antisense probes to pull down the target gene and complexes bound to it, including RNA, proteins, and even DNA. RNA-protein pulldown uses probes with the sense sequence of the target gene to fish for binding proteins.
2)
2) RAP is essentially nucleic acid hybridization between antisense probes and the target gene RNA, similar to FISH, with reaction temperatures generally around 37°C. RNA-protein pulldown is essentially binding interactions between long nucleic acid probes and proteins, with reaction temperatures at 4°C.