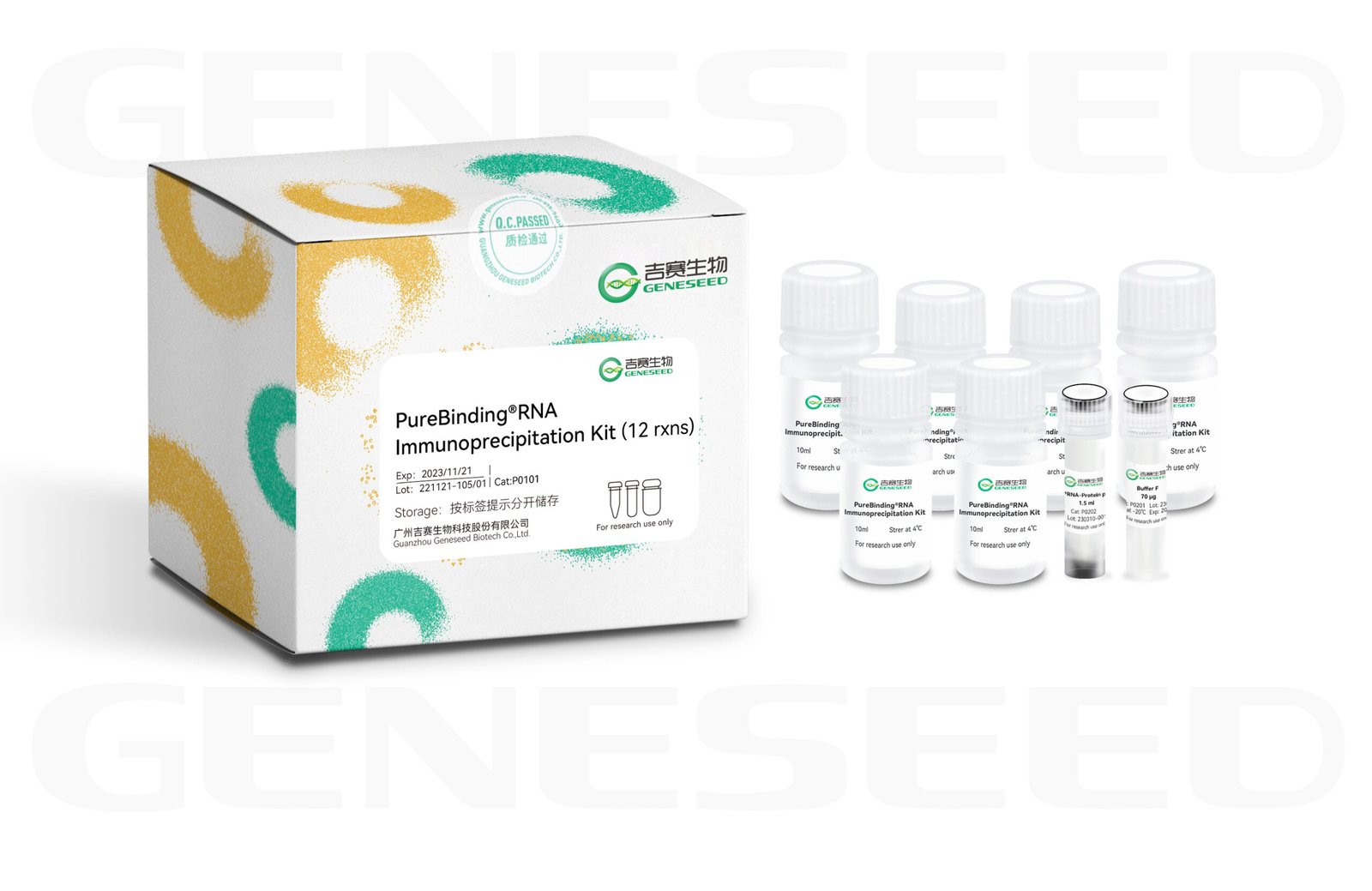
RNA Immunoprecipitation (RIP) is an experimental technique for studying intracellular RNA–protein interactions. Using an antibody against the target protein, the target protein and the RNAs bound to it are precipitated; after separation and purification, the RNAs in the complex can be analyzed by Microarray (RIP-chip), quantitative qPCR, or high-throughput sequencing (RIP-Seq). This reagent set includes the complete reagents required for RIP experiments as well as reagents for extraction of RIP products, making the experiment easier and faster.
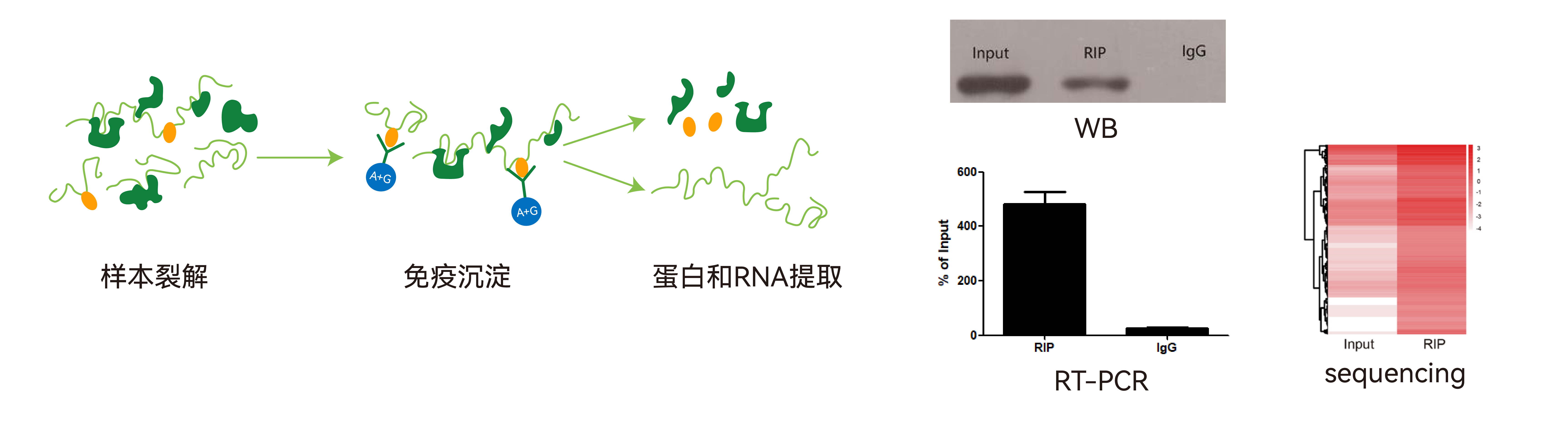
RIP technique schematic
Product Features
1. Convenient and fast: the kit contains complete components, including the full set of reagents for RIP and extraction, and can be completed in 5 hours.
2. System stability: high capture efficiency, stable experimental system, and high reproducibility.
Scope of application
1. Analysis of RNAs and other interacting proteins that bind to the target protein;
2. Identification of RBP-bound RNA motifs;
3. RIP experiments are not suitable for membrane-bound proteins or DNA-binding proteins (histones, nucleolar proteins, etc.).
Experimental procedure
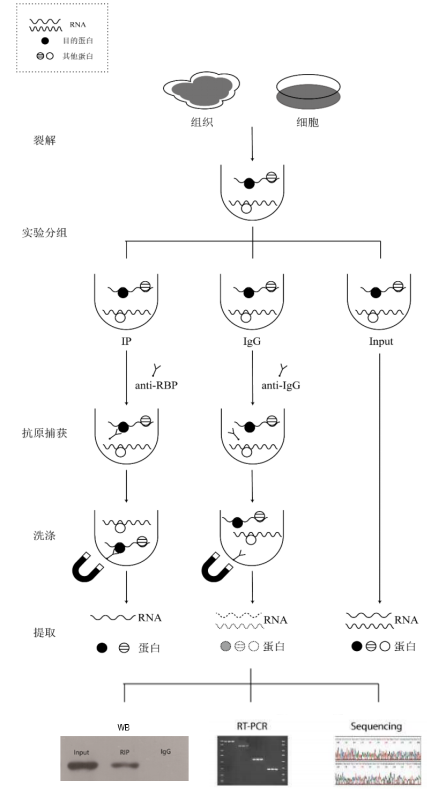
Case Analysis
The purpose of the RIP assay is to analyze the RNAs bound to the target protein. The principle is to use an antibody against the target protein to precipitate the corresponding RNA–protein complexes, then separate and purify them and perform sequencing or PCR analysis on the RNAs associated with the complexes.
On September 23, 2016, Professor Maria R. Matarazzo from Italy introduced the classic RIP assay protocol in the journal Methods in Molecular Biology. The RIP assay workflow described in the article is shown in the figure below:

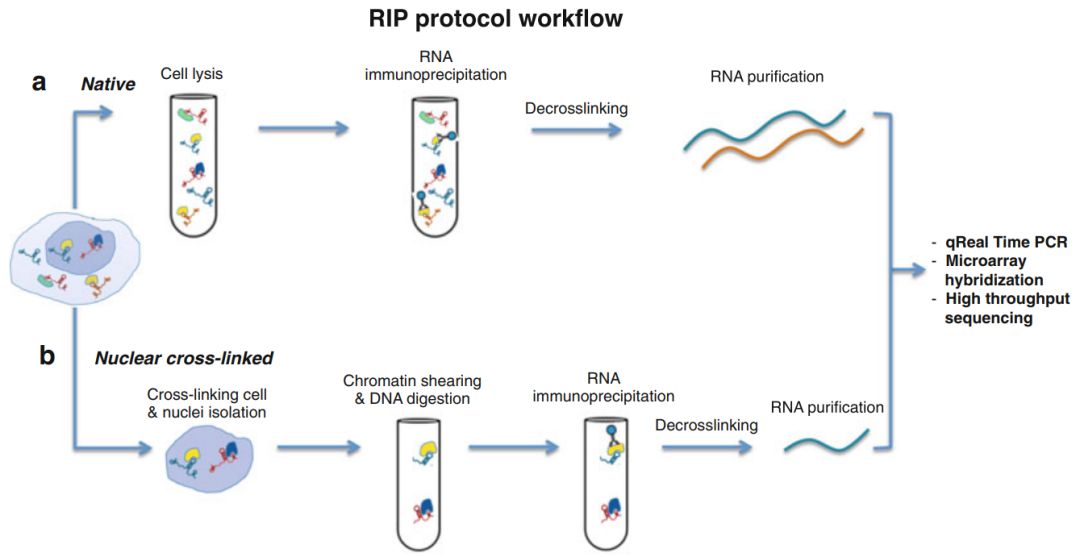
Classic RIP assay workflow (from [1])
What are the applications of the RIP assay? It is well known that RNA is an unstable biological macromolecule; the vast majority of RNAs require formation of RNA–protein complexes with specific RNA-binding proteins to exist stably in cells. RNA–protein complexes drive post-transcriptional regulation of gene expression in virtually all cellular processes, including splicing, nuclear export, mRNA stability, and translation. Therefore, understanding gene regulation depends on identifying changes in RNA binding in these processes. RIP technology was developed for this purpose and can be applied to studies of miRNA regulatory targets, interactions between RNA and RNPs (RNA-binding proteins), and so on.
Case1
RIP for studying circRNA–protein interactions
On March 29, 2022, Professor Chen Junxia from Chongqing Medical University published an article in Mol Cancer titled “Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10.” This study revealed a novel mechanism whereby hypoxia-induced circWSB1 can interact with USP10, thereby attenuating USP10-mediated p53 stabilization and promoting BC progression, providing an alternative prognostic biomarker and therapeutic target for BC.

It has been reported that circular RNAs participate in hypoxia regulation via the miRNA sponge mechanism.
Nearly all of these hypoxia-associated circRNAs act as miRNA sponges; circular RNAs can also function by binding proteins and by translating peptides. The authors used pull-down to analyze the binding products of the circular RNA; although P53 was not identified, the authors found that the deubiquitinating enzyme USP10 binds to circWSB1. They predicted the binding sites between the two using databases. Full-length and deletion mutants of USP10 were designed and tagged with Flag (Figure 3); the interaction between USP10 and circWSB1 was validated by RIP experiments (using Gisea Bio RIP kit)
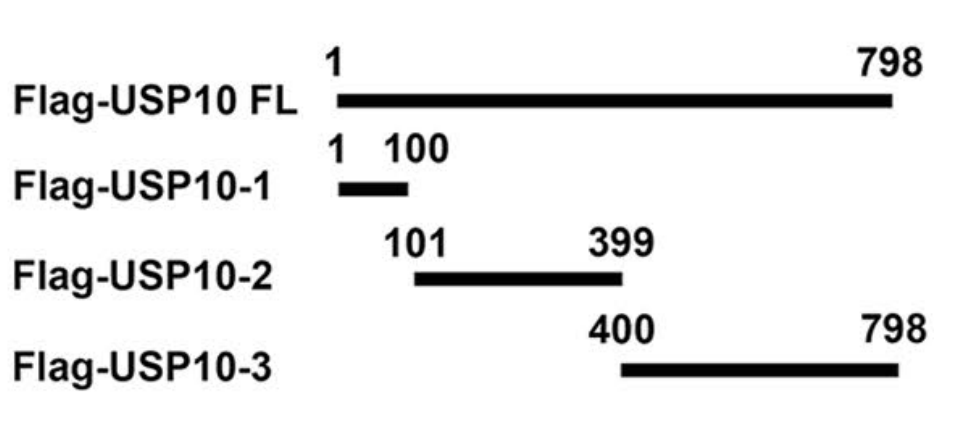
Case2
RIP is used to study lncRNA–protein interactions

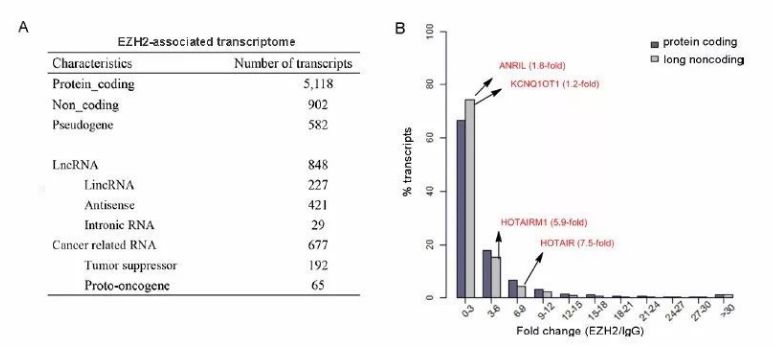
Comparison of the binding capacities of lncRNAs and coding RNAs with EZH2 (from [3])
In this study, the following were used
EZH2-specific antibody was used for RIP-seq in human gastric cancer cell lines MKN45 and AGS, and in control cell line GES-1, analyzing 8,256 EZH2-associated RNAs, including coding and noncoding RNAs, among which MALAT1 was highly expressed in the MKN45 cell line. It was further found that MALAT1 interacts with EZH2 to suppress the expression of protocadherin 10 (PCDH10), thereby promoting invasion and metastasis of gastric cancer cells [3].
Case3
RIP for studying miRNA–protein interactions

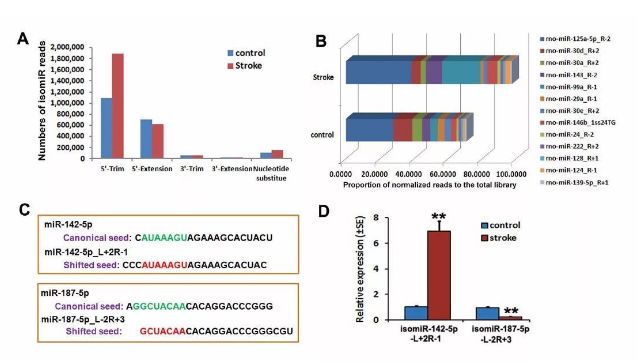
isomiR analysis in iron-deficient and non-ischemic NPCs (from [4])
In this study, using
Ago2 RIP-seq was used to analyze the expression of miRNAs in neural progenitor cells (NPCs) from the subventricular zone (SVZ) of adult nonischemic and ischemic rats [4].
Results show that, compared with non-ischemic
NPCs, stroke altered the expression of Ago2-associated miRNAs in NPCs; the number of isomiR reads increased in ischemic NPCs, with the isomiR-142-5p_L+2R-1 (with 5′ end nucleotide additions) significantly upregulated and the isomiR-187-5p_L-2R+3 (with 5′ end nucleotide deletions) significantly downregulated [4].
Case analysis references
[1] RIP: RNA Immunoprecipitation. Methods Mol Biol, 2016. 1480: p. 73-86.
[2] Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10[J]. Molecular Cancer, 2022, 21(1):1-20.
[3] MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget. 2016 Mar 15; 7(11): 12693–12703.
[4] Identification of miRNomes associated with adult neurogenesis after stroke using Argonaute 2-based RNA sequencing. RNA Biol. 2017; 14(5): 488–499.
Customer Article
[1] IF27.401 The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. Molecular Cancer
[2] IF17.388 N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. Journal of Hematology & Oncology
[3] IF14.919 The genomic landscape of cholangiocarcinoma reveals the disruption of post-transcriptional modifiers. Nature Communications
[4] IF11.161 A novel lncRNA ARST represses glioma progression by inhibiting ALDOA-mediated actin cytoskeleton integrity. JOURNAL OF EXPERIMENTAL & CLINICAL CANCER RESEARCH
[5] IF9.261 Mettl14 mediates the inflammatory response of macrophages in atherosclerosis through the NF-κB/IL-6 signaling pathway. CELLULAR AND MOLECULAR LIFE SCIENCES
[6] IF8.886 ac4C acetylation of RUNX2 catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents ovariectomy-induced bone loss. Molecular Therapy-Nucleic Acids
[7] IF8.718 The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. EXPERIMENTAL AND MOLECULAR MEDICINE
[8] IF8.679 IGF2BP2-dependent activation of ERBB2 signaling contributes to acquired resistance to tyrosine kinase inhibitor in differentiation therapy of radioiodine-refractory papillary thyroid cancer. CANCER LETTERS
[9] IF8.469
A novel 3’ tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer. Cell Death & Disease
[10] IF8.469 SF3B4 promotes ovarian cancer progression by regulating alternative splicing of RAD52. Cell Death & Disease
[11] IF8.469 Linc-ROR facilitates progression and angiogenesis of hepatocellular carcinoma by modulating DEPDC1 expression. Cell Death & Disease
[12] IF8.469 A new candidate oncogenic lncRNA derived from pseudogene WFDC21P promotes tumor progression in gastric cancer. Cell Death & Disease
[13] IF8.469
HNRNPA1-mediated exosomal sorting of miR-483-5p out of renal tubular epithelial cells promotes the progression of diabetic nephropathy–induced renal interstitial fibrosis. Cell Death & Disease
Q & A
1. Can RIP be performed for membrane proteins or nuclear proteins?
In order not to disrupt interactions between endogenous proteins and
RNA, the RIP lysis buffer acts relatively gently. Experimental evidence has shown that the RIP lysis buffer can lyse the plasma membrane and nuclear envelope, releasing cytoplasmic and nuclear proteins, but proteins that are tightly associated with DNA or cell membranes cannot be lysed and released; therefore it is not suitable for RIP of DNA- or membrane-bound proteins.
2. Why is the RNA concentration obtained from the RIP kit very low—is that normal?
The yield from RIP is generally insufficient to directly measure concentration. Analysis can be performed by quantitative RT-PCR (if the RBP’s binding target genes are known) or by sequencing technologies.
For qPCR analysis, because the RNA concentration is unknown, you may use the maximum loading volume of the reverse transcription system for the reverse transcription. For sequencing, if the yield from a single experiment is insufficient for library construction, you may repeat the experiment and pool the products, or appropriately increase the initial sample amount.
3. Is the RIP kit RNA product analysis quantitative or semi-quantitative?
It is a semi-quantitative analysis, to
Determine whether there is an interaction based on the fold change in relative expression between the Ip and IgG groups.
4. How is the 5 µg antibody stated in the RIP manual calculated?
If the antibody datasheet provides a mass concentration, you can convert accordingly. If not, you can use the volume recommended in the datasheet.
5. In RIP experiments, do the antibodies used for IP and WB need to be from different species?
在
In RIP experiments, it is preferable that the antibodies used for IP and WB come from different species; otherwise, when running WB, strong heavy and light chain bands may appear at ~55 kDa and ~25 kDa, respectively, affecting the exposure of the target protein.
Detailed information can be found at:
How to avoid
Heavy chain and light chain interference during IP detection_China Bio-Equipment Network (bio-equip.com)
How to avoid immunoprecipitation co-precipitation
IgG cross-contamination in Co-immunoprecipitation (Co-IP) – Baidu Wenku (baidu.com)
6. Is there an alternative to acetone for the protein extraction step in RIP?
Acetone is suitable for the experimental procedures described in this kit manual. Customers may also use other methods for protein precipitation and extraction.
7. After adding β-mercaptoethanol to Buffer E, white flocculent precipitate appears — is this normal?
Normal. Let the sample equilibrate at room temperature for a short time; even if white precipitate remains, it does not affect use.
8. RIP results show normal WB detection, and the Ct values for the IP and IgG groups are almost identical—what does this indicate?
It indicates that the target protein does not bind specifically to the target gene.
9. Is the RIP kit suitable for lysing bacteria?
Bacterial samples should be snap-frozen in liquid nitrogen and ground, then proceed with these lysis steps.
10. Can MeRIP and RIP techniques or kits be used interchangeably?
Although MeRIP and RIP share the same core principle, their experimental methods, workflows, antibodies used, and applications are different and they are not interchangeable.
Learn more:
https://mp.weixin.qq.com/s/S84RXllkC0CegN61EZ84hA
https://mp.weixin.qq.com/s/520lOrEJrqZV7XutP4KE_Q