Transcription is the key step in the transfer of genetic information fromDNA to RNA, and it is the primary form of RNA biosynthesis. Transcription factors are proteins that can bind specifically to sequences at the 5’ end of genes, enabling target genes to be expressed with specific strength in specific times and locations; they are one of the key factors involved in transcriptional regulation.
Chromatin immunoprecipitation (Chromatin immunoprecipitation, ChIP) is a method for analyzing DNA–protein interactions. Its basic principle is that live cells or fresh tissues are crosslinked to lock nuclear proteins (transcription factors, DNA damage repair proteins, histones or DNA modification enzymes, reader enzymes, eraser enzymes, etc.) in interaction with target genes; chromatin is then fragmented by sonication or enzymatic methods, and finally DNA fragments bound by nuclear proteins are enriched by immunoprecipitation using specific antibodies against the nuclear proteins.Nuclear protein–boundDNA fragmentsafter isolation and purification,can be analyzed by quantitativeqPCR or high-throughput sequencing, etc.
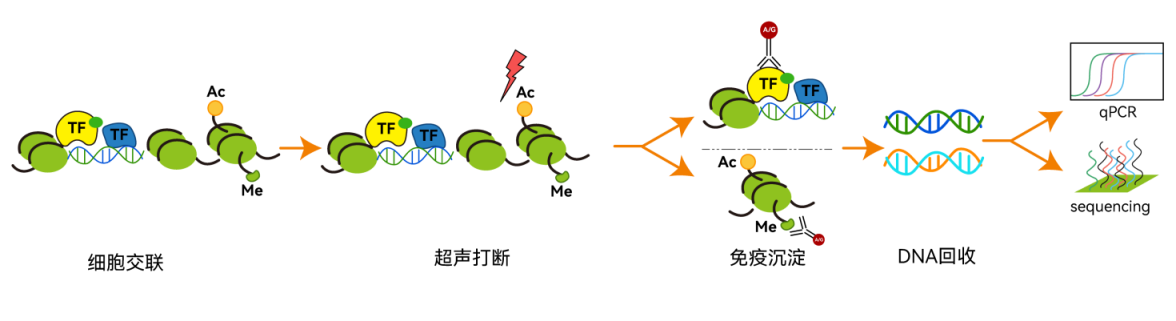
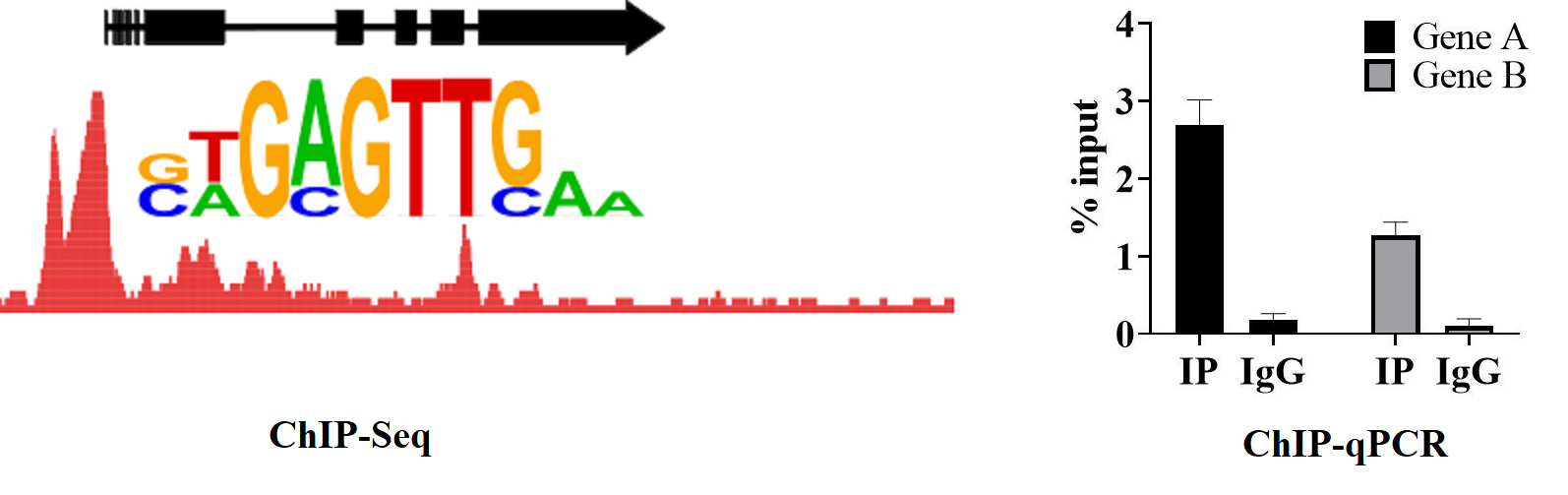
ChIPtechnical principle diagram
Scope of application
1. Validate known protein–DNA interactions;
2. Study the target genes and binding sites of transcription factors;
3. Study the genomic action sites of DNA/RNA polymerases and transcription complexes;
4. Study the relationship between covalent histone modifications and gene expression.
Product advantages
① Intracellular reaction: use antibodies to immunoprecipitate intracellular protein–DNA complexes, truly reflecting the physiological state of the cell;
② Simple procedure: easy to operate;
③ Short time cycle: the entire experimental process<8h.
Case Analysis
Q & A
Question:
After the ChIP experiment is completed, the CT values between the IP and IgG groups show little difference — what is the reason?
Answer:IP group CT value在
A CT value of around 28 may be due to one or more of the following reasons: 1) insufficient sample amount; 2) the detected gene is not a transcription factor target gene;
③ Low antibody titer; ④ Antibody not ChIP-grade; ⑤ Primer design issues.
Question:Is a sample size of 10e6 sufficient to perform ChIP experiments?
Answer: Not recommended. Due to the low probability of transcription factors undergoing transcriptional activation at specific time points, it is necessary toExperiments can only be carried out with 1×10^8 cells.
Question:
How should positive and negative controls for ChIP experiments be chosen, and are positive and negative controls required for the experiment?
Answer: For a negative target gene, you can choose
GAPDH or ACTB, but positive target genes are influenced by cell type, physiological state, drug interventions, and other factors, so they cannot be standardized; consult the literature for recommendations. It is recommended to include negative and positive control detections, but they are not mandatory。
Question:What is the typical concentration range of ChIP product?
Answer:
The total amount and concentration of ChIP products are influenced by many factors such as transcription factor expression levels, the number of target genes, and cellular state, so there is no uniform standard; generally the total amount ranges from 1 to 200 ng。
Question:
If the concentration of ChIP products is low and an ultrasensitive spectrophotometer cannot detect it, does that mean the experiment failed?
Answer:
The total yield and concentration of ChIP products are affected by many factors such as transcription factor expression level, number of target genes, and cell state; the total yield is generally low. The detection range of microvolume spectrophotometers is 10 ~ 1000 ng; values outside this range cannot be detected. qPCR is recommended as the standard for detection.。
Question: None
Can ChIP-grade antibodies and IP-grade antibodies be used to perform ChIP experiments?
Answer:Theory of IP-grade antibodies上Can be performed
ChIP experiments; it is recommended to first validate by CoIP before the experiment to confirm the antibody works;
Question: What are the critical pieces of equipment? Is the sonication step necessary? Are there sonicators with probes and probe-free sonicators such as
Which to choose?
A: The genome must be sonicated before chromatin immunoprecipitation can be performed. The main difference between sonicators with probes and those without probes is operating power; the former has substantially higher power than the latter. The recommended operating power is500 ~ 900 range。
Q:How to choose between CUT&Tag and ChIP techniques?
Answer: Both techniques are used for studying transcription factor target genes. The differences are:
1) Method of genome fragmentation: the former (CUT&Tag) uses enzyme digestion for fragmentation, while the latter (ChIP) uses sonication. If you do not have a sonicator, you can choose the former; 2) In CUT&Tag, fragmentation and library preparation are performed simultaneously, making the procedure simpler; 3) In ChIP experiments, the antibody–transcription factor complexes are larger, leading to greater nonspecificity.较低。
Question:Is it normal for the Ct value of the input sample qPCR to be above 28?
Answer: Not normal,
For a sample amount of 10e7 cells, the standard experimental Ct value is in the range of 20–24. If it exceeds this range, possible reasons are: ① insufficient cell number; ② low cell viability; ③ failed recovery; the experiment needs to be repeated.。
Question: In the step of beads binding antibody, is it normal that the mixture does not flow when shaken vertically?
A: Normal. To improve antibody–bead binding efficiency, a smaller binding volume is required; although no bulk solution movement is visible to the naked eye during shaking, the magnetic beads are continuously suspended and do not sediment.
Download
Gisai Bio PureBinding® Chromatin Immunoprecipitation Kit Cat. No. P0301 P0302.pdf