RNase R is an essential enzymatic tool for circRNA identification and enrichment experiments; it can digest linear RNA to enrich circular RNAs (circRNAs) or lariat RNAs.
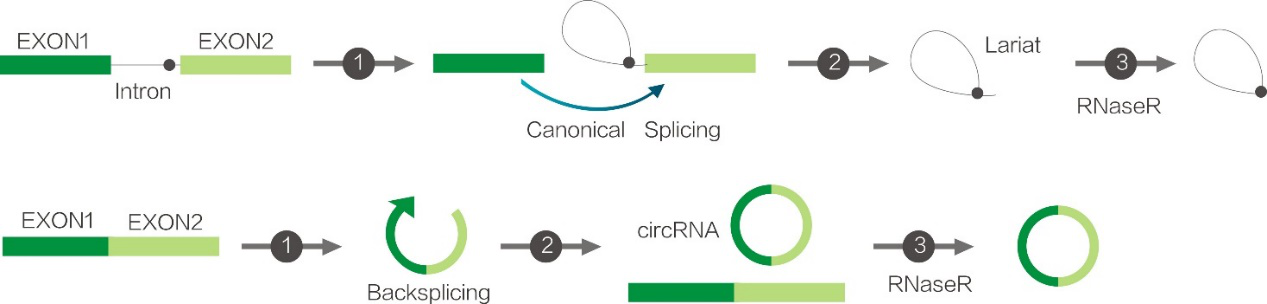
Figure1. Schematic of RNase R digestion of RNA
Applications
1. Identification of circRNA
Determinewhether the tested molecule is circRNA based on whether a band is detected in the RNase R(+) and RNase R(-) groups.
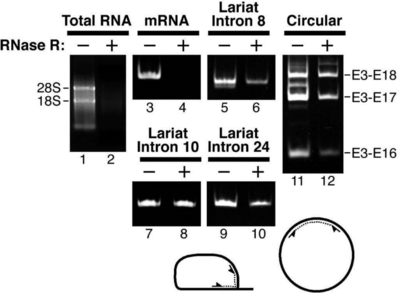
Figure2. RT-PCR detection of Lariat/Circular RNA after RNase R digestion
In Figure2, mRNA is detected as a band in RNase R(-) but not in RNase R(+); detection of Lariat/Circular RNA shows bands in both RNase R(-) and RNase R(+) samples, indicating that mRNA is digested while Lariat/Circular RNA is resistant to digestion (Suzuki H et al., 2006).
2. Enrichment of circRNA
High-throughput sequencing often requires enrichment ofcircRNA. To investigate the degree of circRNA enrichment and changes in related genes after RNase R digestion, sequencing can be performed on both RNase R(+) and RNase R(-) sample groups.
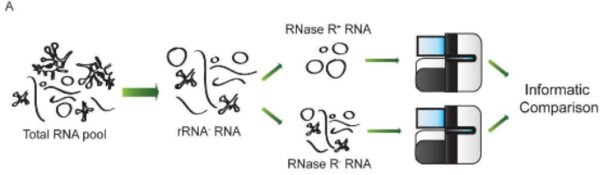
Figure3. Schematic of RNase R(+) and RNase R(-) RNA sequencing (Jeck WR et al., 2014)
Studies have reported that sequencing showsJunction Reads in the RNase R(+) group are enriched 5–10 fold compared with RNase R(-) samples, allowing identification of several thousand to tens of thousands of circRNAs (Figure 3).
3. Purification of circRNA
Eliminating interference from residual linearRNA is key to the synthetic production of high-purity circRNA. Efficient RNase R is not only a critical tool for ligase-based synthetic circRNA methods but is also indispensable for synthetic circRNA produced via intron self-splicing.
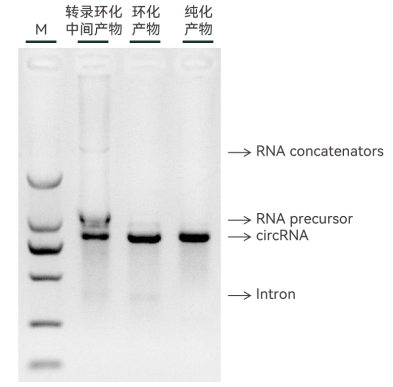
Figure 4. Gisay BiocircPrecise™ platform prepared circular RNA products (“purified products” were purified using GSPure® RNase R)
Product advantages
High specificity: specifically digests linearRNA;
Highly efficient and rapid:most linear RNA can be digested within 5–15 min;
Buffer compatible: digestion products can be used directly in downstream experiments;
Easy to use: simple system,one-step reaction at 37°C.
Quality control
SDS-PAGE purity >95%; total RNA was digested with RNase R and then analyzed by RT-qPCR, showing a marked decrease in linear RNA abundance while circular RNA abundance remained essentially unchanged.
Performance comparison
1.RNA electrophoresis analysis
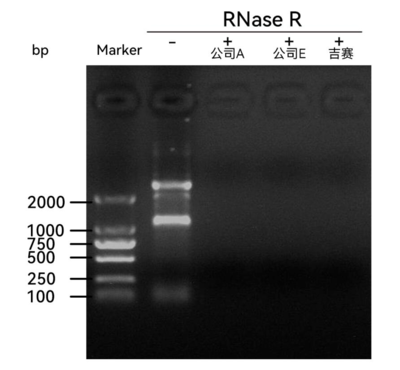
Add 10 U RNase R to 2.5 μg Total RNA and incubate at 37°C for 15 min, then proceed directly to electrophoresis. The results show that the bands in the RNase R(+) group are reduced (not visible), indicating that RNase R digests Total RNA. The RNase R from Gesei and Company A or Company E exhibits comparable digestion effects on Total RNA.
3.RT-qPCR assay
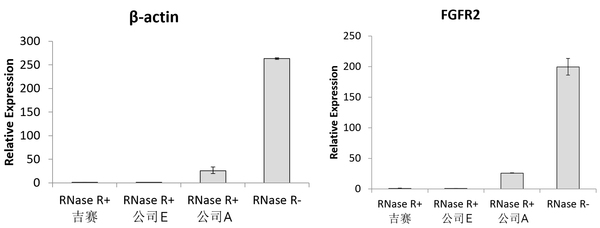
Add10 U RNase R to 2.5 μg total RNA, incubate at 37°C for 30 min, and then perform RT-qPCR. Results showed that the abundances of β-actin and FGFR2 were markedly reduced in the RNase R+ group, indicating that RNase R can digest linear RNA. RNase R from Gisen and Company E had comparable effectiveness in digesting linear RNA and outperformed RNase R from Company A.
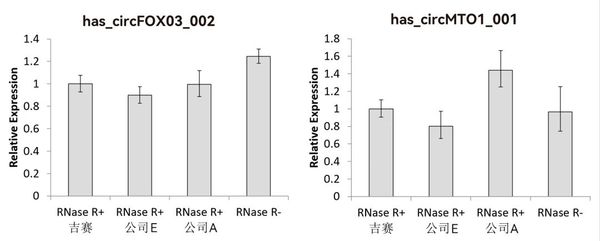
10U RNase R was added to 2.5 μg total RNA and incubated at 37℃ for 30 min before RT-qPCR. The results showed that the abundances of hsa_circFOXO3_002 and hsa_circMTO1_001 in the RNase R+ group remained essentially unchanged, indicating that circRNAs are resistant to RNase R digestion. The RNase R performance from Gesei is comparable to that of Company E or Company A.
Note: Data were calculated using the “RNase R+ Gesei” group as the control, setting its relative abundance value to 1.
Case Analysis
Case1:

On July 21, 2022, Professor Guan Feng’s team from the School of Life Sciences, Northwest University, made new progress in the study of the biological function and regulatory mechanism of C1GALT1 in bladder cancer. The research results were published in Journal of Experimental & Clinical Cancer Research, entitled “Dysregulation and prometastatic function of glycosyltransferase C1GALT1 modulated by cHP1BP3/ miR-1-3p axis in bladder cancer.”
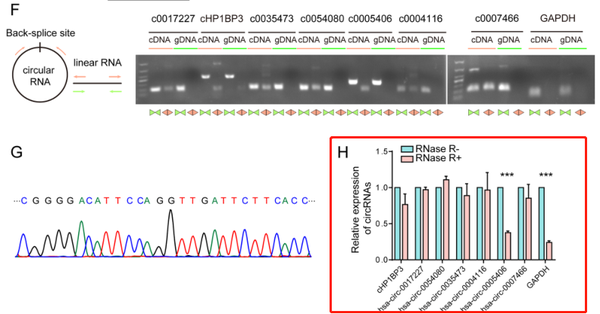
The research team used
RNase R (R0301, Genesea Biotech) to assess the sensitivity of seven DEcircRs to RNase R digestion, in order to exclude back-splicing potentially produced by trans-splicing, genomic rearrangement, or PCR artifacts. It was demonstrated that the resistance of the DEcircRs to RNase R was higher than the GAPDH control.
Case2:

On August 12, 2022, the research team led by Prof. Wei-Guang Zhang of the School of Basic Medical Sciences, Peking University published online in the journal Redox Biology a research paper titled “CircSV2b participates in oxidative stress regulation through miR-5107-5p-Foxk1-Akt1 axis in Parkinson’s disease.” The study found that the circRNA molecule circSV2b plays an important role in the pathogenesis of Parkinson’s disease (PD) and elucidated its mechanism of action.
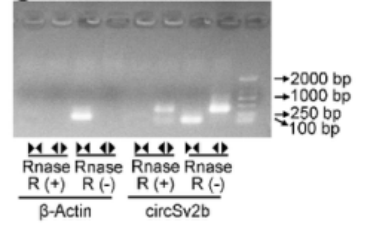
The research team used
RNase R (R0301, GENESESEA) treatment markedly reduced the mRNA levels of linear SV2b and β-Actin, while no significant change was observed in the amount of circSV2b, demonstrating that circSV2b is a highly stable, intron-retaining circular RNA.
Customer Article
[1] IF27.401 A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Molecular Cancer
[2] IF15.302 circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Molecular Cancer
[3] IF15.302 Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Molecular Cancer
[4] IF13.493 The circFASN/miR-33a pathway participates in tacrolimus-induced dysregulation of hepatic triglyceride homeostasis. Signal Transduction and Targeted Therapy
[5] IF12.658
Dysregulation and prometastatic function of glycosyltransferase C1GALT1 modulated by cHP1BP3/miR-1-3p axis in bladder cancer.JOURNAL OF EXPERIMENTAL & CLINICAL CANCER RESEARCH
[6] IF11.556 Engineering circular RNA regulators to specifically promote circular RNA production. Theranostics
[7] IF11.492 CircIMMP2L promotes esophageal squamous cell carcinoma malignant progression via CtBP1 nuclear retention dependent epigenetic modification. Clinical and Translational Medicine
[8] IF11.161
Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma.JOURNAL OF EXPERIMENTAL & CLINICAL CANCER RESEARCH
[9] IF11.161
Warburg effect–promoted exosomal circ_0072083 release up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma.JOURNAL OF EXPERIMENTAL & CLINICAL CANCER RESEARCH
[10] IF11.161 CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated β-catenin stabilization. JOURNAL OF EXPERIMENTAL & CLINICAL CANCER RESEARCH
[11] IF11.161 CircAGAP1 promotes tumor progression by sponging miR-15-5p in clear cell renal cell carcinoma.
JOURNAL OF EXPERIMENTAL & CLINICAL CANCER RESEARCH
[12] IF10.392 A novel intronic circular RNA, circGNG7, inhibits head and neck squamous cell carcinoma progression by blocking the phosphorylation of heat shock protein 27 at Ser78 and Ser82. Cancer Communications
[13] IF10.122
Lipotoxicity-induced circGlis3 impairs beta cell function and is transmitted by exosomes to promote islet endothelial cell dysfunction.DIABETOLOGIA
[14] IF8.886 circRNA-0002109 promotes glioma malignant progression via modulating the miR-129-5P/EMP2 axis.
Molecular Therapy – Nucleic Acids
[15] IF8.886 HnRNP-L-regulated circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes progression in prostate cancer.
Molecular Therapy – Nucleic Acids
[16] IF8.886
Effects of long-term culture on the biological characteristics and RNA profiles of human bone marrow-derived mesenchymal stem cells.
Molecular Therapy – Nucleic Acids
[17] IF8.886 CircSTK40 contributes to recurrent implantation failure via modulating the HSP90/AKT/FOXO1 axis.
Molecular Therapy – Nucleic Acids
[18] IF8.579 circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis. Theranostics
[19] IF8.469
Circular RNA hsa_circ_0043280 inhibits cervical cancer tumor growth and metastasis via the miR-203a-3p/PAQR3 axis.Cell Death & Disease
[20] IF8.469 CircHAS2 promotes the proliferation, migration, and invasion of gastric cancer cells by regulating PPM1E mediated by hsa-miR-944. Cell Death & Disease
[21] IF8.469 Autophagy-related circRNA evaluation reveals hsa_circ_0001747 as a potential favorable prognostic factor for biochemical recurrence in patients with prostate cancer. Cell Death & Disease
[22] IF8.469 Circular RNA circACSL1 aggravated myocardial inflammation and myocardial injury by sponging miR-8055 and regulating MAPK14 expression. Cell Death & Disease
[23] IF8.469 Circular RNA circRUNX1 promotes papillary thyroid cancer progression and metastasis by sponging MiR-296-3p and regulating DDHD2 expression. Cell Death & Disease
Q & A
1. What are the criteria for successful RNase R digestion?
Recommendations
Take a portion of the RNA after RNase R digestion and run it directly on a gel; attenuation of the 28S and 18S bands indicates successful digestion. (Gel results are straightforward and can directly show digestion success; if digestion fails, that indicates a problem.)
2. How should control groups be designed for an RNase R experiment?
Refer to the figure below for multiple groupings and controls to rule out influencing factors. (Adding multiple controls can help exclude and identify the cause of any abnormalities.)
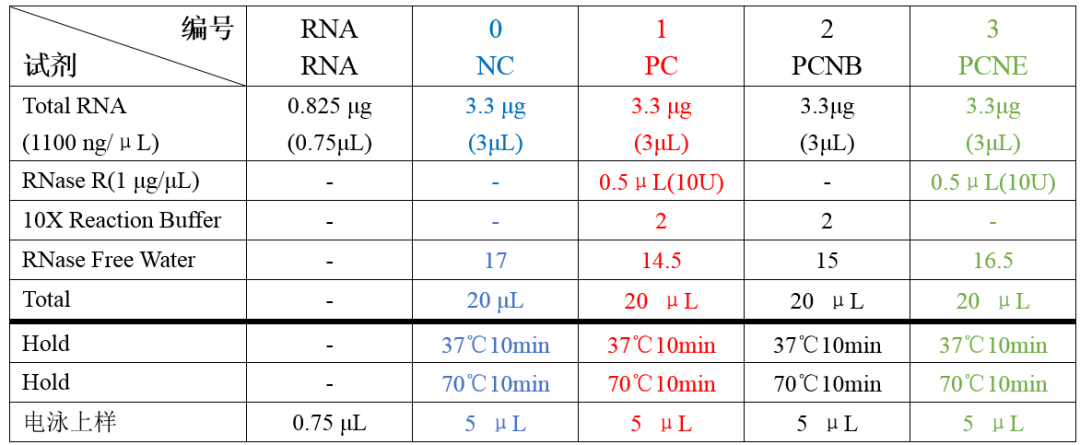
3. What are the conditions for RNase R experiments?
Digestion time and
The amount of RNase R may require preliminary experiments to determine the appropriate conditions so that linear RNA is digested while circRNA remains unchanged. Generally, digestion at 37°C for 15 min, with the Ct value of internal controls delayed by 5–10 cycles, is considered a relatively successful result. (If digestion is insufficient or excessive, or if the resulting values are unsatisfactory, preliminary experiments can be performed to optimize conditions.)
4. What are the issues with RNase R primer testing?
Pay attention to the specificity of the detection primers. Be careful to distinguish
homologous sequences between circRNA and mRNA. (When primers produce nonspecific amplification, the measured results are completely unreliable; any claim that digestion had no effect or had excessive effect is not credible.)
5. If the amount of circRNA does not change, is it considered a success?
总
After RNA digestion, the amount of linear RNA decreases while the amount of circRNA remains essentially unchanged, so the relative proportion of circRNA increases — that is, circRNA is enriched. Subsequent qPCR measurements of circRNA abundance may therefore be biased upwards, which is normal.
6. How to recover RNA after RNase R digestion?
If the digestion solution after RNase R digestion needs to be recovered, for convenience Trizol can be used for recovery; follow the manufacturer’s instructions. If you do not recover and proceed directly to reverse transcription, the volume of digestion solution taken up must not exceed one-quarter of the reverse transcription system, because residual ions and other components in the digestion solution can interfere with downstream RT-qPCR reactions. For example, if the reverse transcription is a 20 µl system, the RNase R digestion solution taken should not exceed 5 µl.
7. What to do if RNase R digestion is unsuccessful?
Re-extract using a new kit or fresh reagents
RNA, ensure the purity and quality of RNA. (High RNA purity is required for efficient digestion; residual phenol and similar contaminants will significantly affect digestion efficiency, and gDNA contamination will also reduce digestion effectiveness)
8. How to store RNase R
For use within the same day,
RNase R (20 U/µl) can be diluted with nuclease-free water; for long-term use, it should be diluted with Storage Buffer, the formula of which is provided in the manual. (When the required amount is small the volume is small; some customers wish to dilute for use)
9. Precautions for RNase R digestion
Preparation
Use nuclease-free reagents; nuclease-free water, pipette tips and other consumables should be used, or water treated with DEPC. In addition, the bench surface (it is recommended to perform the whole procedure in a laminar flow hood or biosafety cabinet) should be sprayed and wiped with nuclease-removal solution. These are standard preparations for RNA experiments.
10. Special situations in RNase R digestion
Special cases: some linearRNA is resistant to digestion, although some circRNAs are also easily digested.
11. After RNase R digestion, is it possible to run the gel directly without purification?
Okay. You can also assess the digestion efficiency.28. The fading of the 18S band indicates successful digestion.
Alternatively, proceed directly
to RT-PCR; generally purification can be omitted, and after enzyme inactivation you can proceed directly to the reverse transcription reaction.
12. How much RNase R is appropriate to use in the digestion reaction?
In different reference articles
The amount of RNase R used and the reaction system vary; the most common is a 20 μL reaction system, using a ratio of 1–4 U RNase R/μg RNA, and optimization may be required in experiments.
13. What are the RNase R digestion reaction temperature and time settings?
RNase R digestion is generally performed at 37°C, and most linear RNA can be digested within 10–30 min. Commonly, 5–15 min digestion is sufficient to obtain very good results.
14. Is RNase R inactivation necessary after the digestion reaction?
For samples in which reverse transcription is performed directly after RNase R digestion, it is recommended to inactivate RNase R by holding at 70°C for 10 min after the 37°C reaction (65°C for 15 min is also used). If purification and recovery are performed after digestion, inactivation may be omitted.
15. RNase R digestion identification
Samples
Take a portion of the sample before and after RNase R digestion and run it directly on a gel to visually assess the quality of the extracted RNA and the effect of RNase R digestion; the fading of the 28S and 18S bands indicates successful digestion. If RNA purity is not high, the 28S/18S bands may show concave trailing. If RNA has gDNA contamination, residual gDNA bands may remain.
16. RNase R digestion reaction conditions?
Different
circRNAs have varying tolerance to RNase R; the specific reaction time needs to be empirically determined. Typical reaction times are 10–30 min; prolonged digestion is not recommended. Some circRNAs are weakly resistant to RNase R digestion. In such cases, digestion time can be reduced.
17. After RNase R digestion, is it necessary to measure the product concentration?
Do not directly measure
RNA concentration, because proteins, salt ions and other components in the reaction system will cause inaccurate RNA concentration measurements. It is recommended to purify the RNA before measuring the concentration.
18. Post-treatment of products after RNase R digestion:
Directly proceed to the reverse transcription reaction is recommended
RNase R inactivation. Inactivation conditions: 70°C for 15 min. The volume of the digestion solution taken up must not exceed 1/4 of the reverse transcription system, because residual ions in the digestion solution can interfere with downstream RT-qPCR reactions. For example, if the reverse transcription is a 20 µl system, the RNase R digestion solution taken must not exceed 5 µl.
If purification and recovery are to be performed after digestion, enzyme inactivation can be omitted.
Download
GSPure® RNase R from GISAID Biotech Cat. No. R0300 R0301 R0302.pdf